f-BGM
Home
Introduction
We proposed a deep learning framework specifically for the fungal genome mining problem, termed as f-BGM. By leveraging a novel self-attention-based pretrained model acquiring the knowledge of inter-Domain Locally Co-occurrent relationship in Fungal genome (f-DLC), f-BGM outperforms existing baselines in both (1) BGC detection and (2) core enzyme identification tasks. In addition, attention weight-based analyses demonstrate that f-BGM is of decent interpretability on deciphering single-domain and -protein importance as well as inter-domain partnership.
Model architecture
The f-DLC architecture
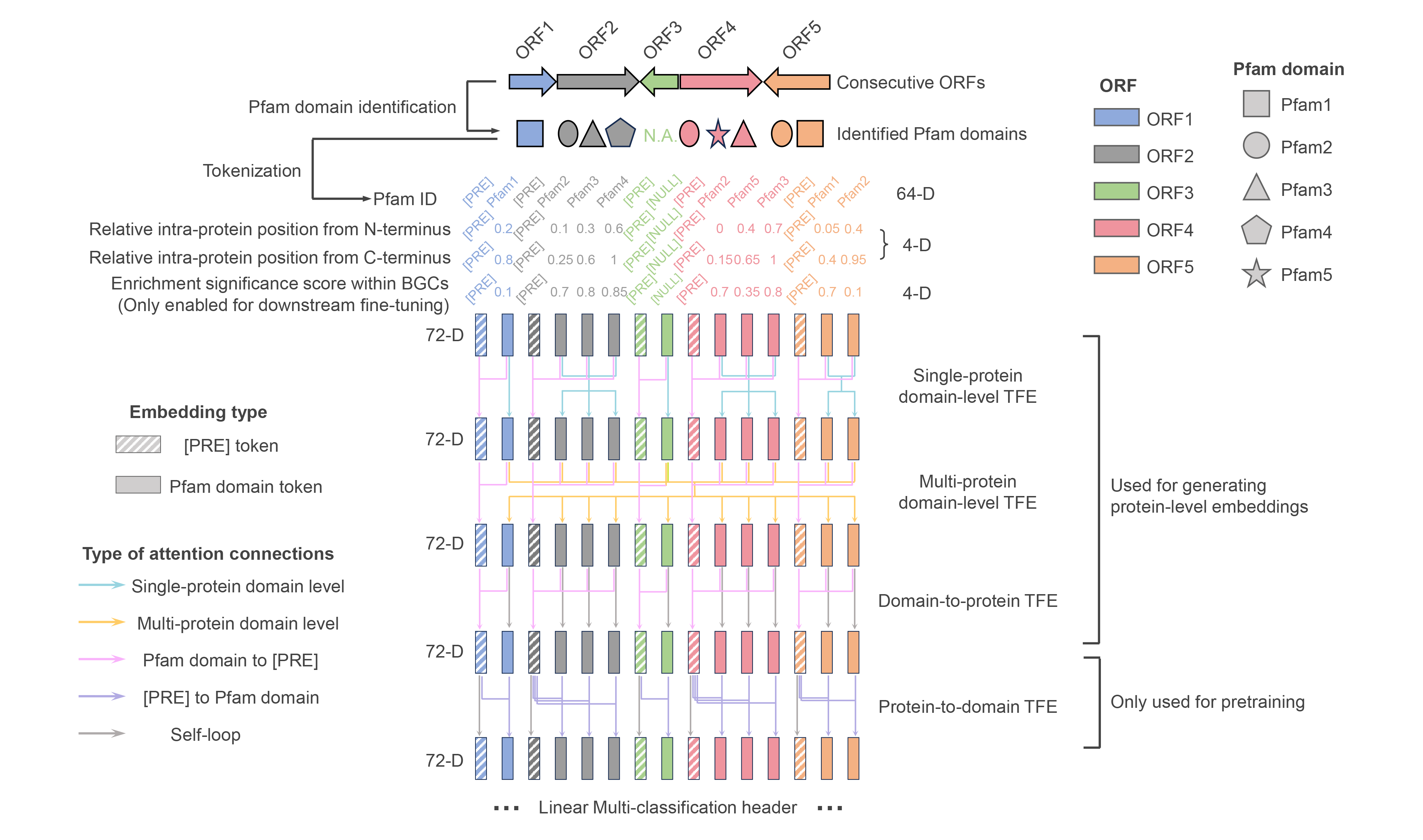
f-DLC receives ≤26 consecutive ORFs as inputs. First, f-DLC transforms the ORFs into linearized token sequences with a hierarchical domain-to-protein structure, which consists of Pfam domain tokens prefixed with [PRE] token for each ORF. Then the tokens are translated into 72-D embeddings consisting of three components: (1) 64-D domain-specific learnable embeddings; (2) 4-D embeddings encoding domains’ relative positioning information in protein; (3) 4-D embeddings encoding domains’ enrichment information in BGC (only enabled during downstream establishment of f-BGM). Next, the embeddings are passed through four sequential transformer encoders (TFE) with different attention masks to achieve layer-wise information interaction. Specifically, the first and second TFE mainly focus on inter-domain information interaction within single protein and multiple proteins, respectively. Meanwhile the attention connections from domain tokens to corresponding prefix tokens are enabled for real-time domain-to-protein information aggregation. The third TFE only retains the domain-to-protein attention mechanism to fully summarize the upstream extracted features and thereby generate meaningful protein-level embeddings for downstream use. The fourth TFE reversely calculates on the attention connections from prefix tokens to domain tokens to satisfy domain-level BERT-style pretraining, where 15% of the input domain tokens are masked and the model is guided to recover these tokens by a downstream multi-classification header.
The f-BGM architecture
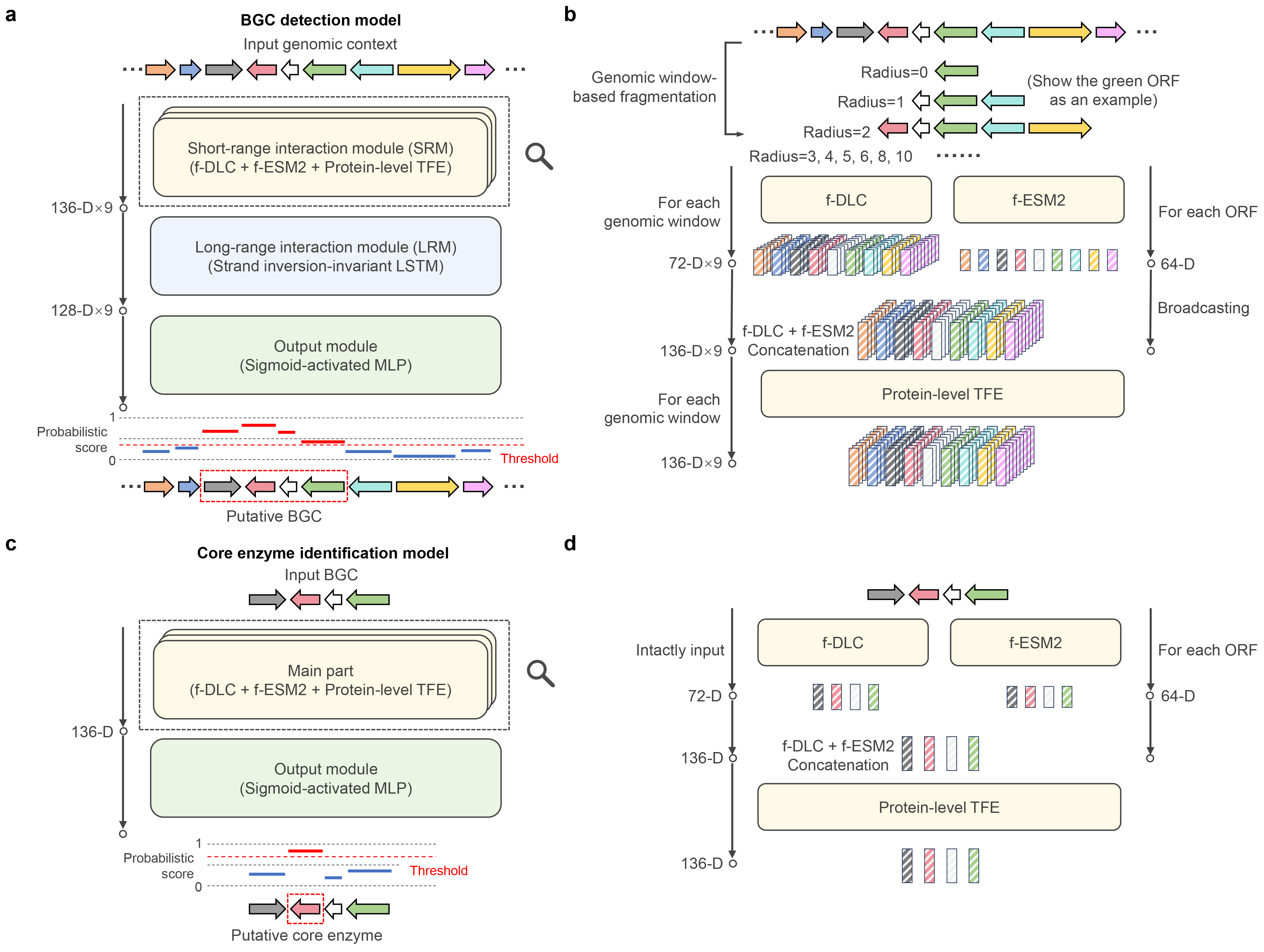
a The BGC detection model receives genomic contexts as input and outputs ORF-level probabilistic scores representing BGC membership. The entire process consists of three sequential steps: (1) self-attention-based short-range information interaction (by SRM), (2) long-short-term memory (LSTM)-based long-range information passing (by LRM) and (3) probabilistic score generation (by OM). b Detailed illustration of the SRM. SRM integrates two pretrained models respectively capturing (1) inter-domain locally co-occurrent relationship in fungal genome (f-DLC) and (2) amino acid patterns within fungal protein sequences (f-ESM2). The input genomic contexts are first fragmented into multiple sizes of genomic windows and fed into f-DLC to enable domain-level information interaction and generate window-wise ORF-level embeddings, meanwhile the ORF-encoded protein sequences are passed into f-ESM2. The f-DLC-output embeddings and broadcasted f-ESM2-output embeddings are further concatenated and fed into an additional transformer encoder for protein-level information interaction (protein-level TFE). c The core enzyme identification model receives BGCs as input and outputs ORF-level probabilistic scores representing the presence or absence of target core enzymes. d The main part of core enzyme identification model mimics the SRM architecture in (b) but without genomic window-based fragmentation on the input BGCs due to their determined borders.
Model performance
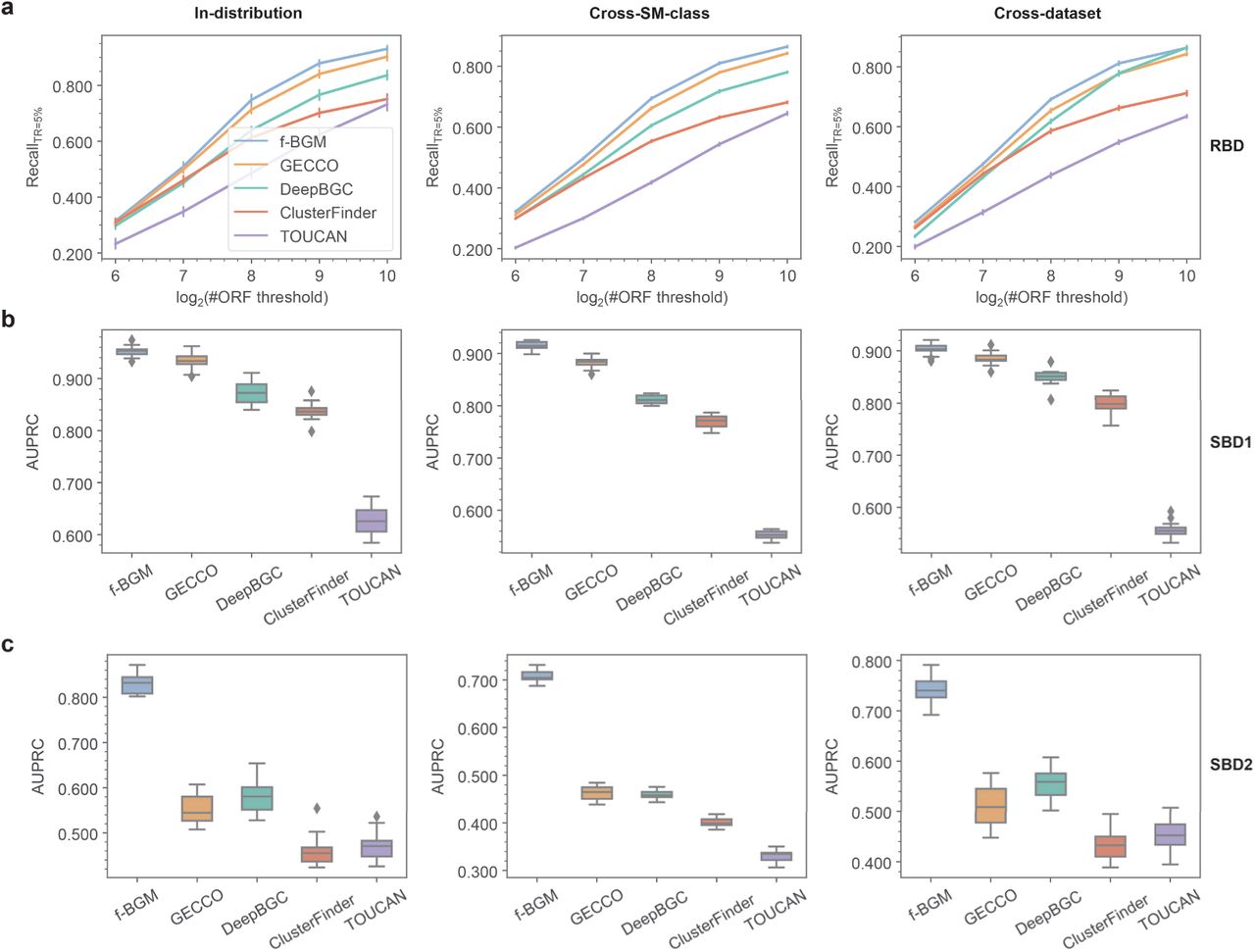
a Performance comparison among f-BGM and four baseline algorithms (ClusterFinder, DeepBGC, GECCO and TOUCAN) with regard to RecallTR=5% in real BGC detection tasks under in-distribution, cross-SM-class and cross-dataset schemes. To avoid biases to overlength contigs, the ORFs included for analyses per contig are limited by a specified maximal number by removing excessive ORFs distant from the known positives. Here the ORF number thresholds of 64, 128, 256, 512 and 1024 are enumerated to cover more situations (x-axis). b, c Performance comparison among f-BGM and four baseline algorithms with regard to AUPRC in (b) standard simulated BGC detection and (c) challenging simulated BGC detection tasks under in-distribution, cross-SM-class and cross-dataset schemes. RBD: real BGC detection; SBD1: standard simulated BGC detection; SBD2: challenging simulated BGC detection.