f-BGM
Tutorial
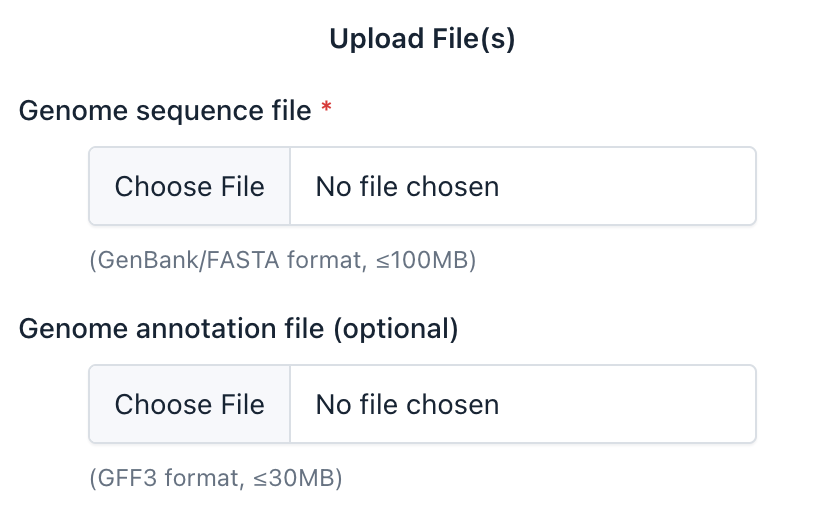
1. Upload File(s)
You must provide a genome sequence file (in GenBank or FASTA format, ≤100MB).
Optionally, you can also upload a genome annotation file (in GFF3 format, ≤30MB).
The annotation file will be used to improve Pfam domain extraction. If omitted, f-BGM will use AUGUSTUS to perform gene prediction on the input sequences.
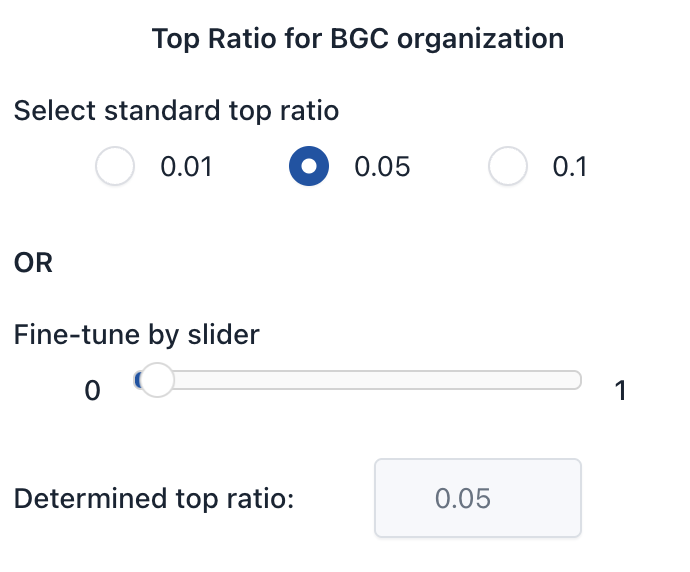
2. Select Top Ratio
Top Ratio determines the top percentage of highly scored ORFs that are considered for BGC organization.
- Select a preset:
0.01,0.05(default), or0.1 - Or use the slider for fine-tuning. The final value will show in the "Determined top ratio" field.
3. Configure Advanced Options
- Min #ORF required for contig(s): Contigs containing fewer than this number of predicted peptides (ORFs) will be excluded from analysis. (default: 32)
- #ORF range of BGC(s): Only predicted BGCs with a number of peptides (ORFs) within this range will be retained. BGCs with fewer than the minimum or more than the maximum will be discarded. (default: 1–128)
- Max #ORF interval for BGC merging: If two candidate BGCs are separated by no more than this number of intergenic ORFs, they will be merged into one. (default: 1)
- Flanking DNA length (bp): When exporting DNA sequences of BGCs, this number of base pairs will be included upstream and downstream. (default: 1000)
- E-value cutoff for Pfam domains identification: Defines the maximum E-value for Pfam domain hits to be considered statistically significant. (default: 0.01)

4. E-mail Notification (available in future version)
Enter your email address to receive notification upon task completion.
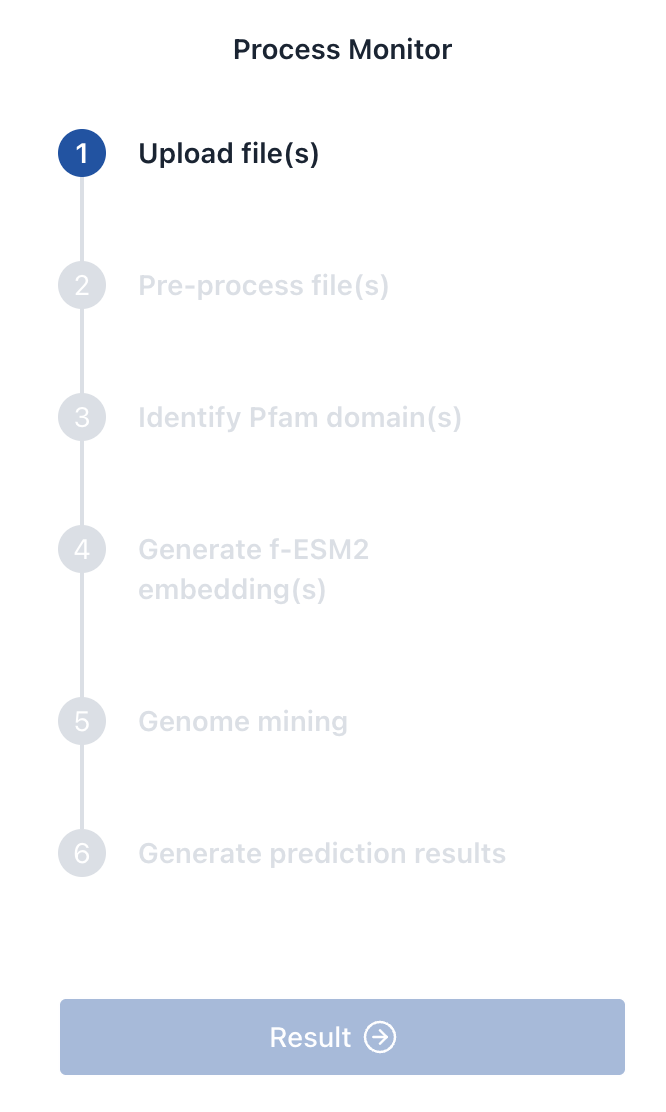
5. Task Execution
Click Submit to launch the genome mining task. You can monitor its progress via the panel. The following steps will be tracked:
- Upload file(s)
- Pre-process file(s)
- Identify Pfam domain(s)
- Generate f-ESM2 embedding(s)
- Genome mining
- Generate prediction results
When all steps are complete, the Result button will become available for downloading prediction files.